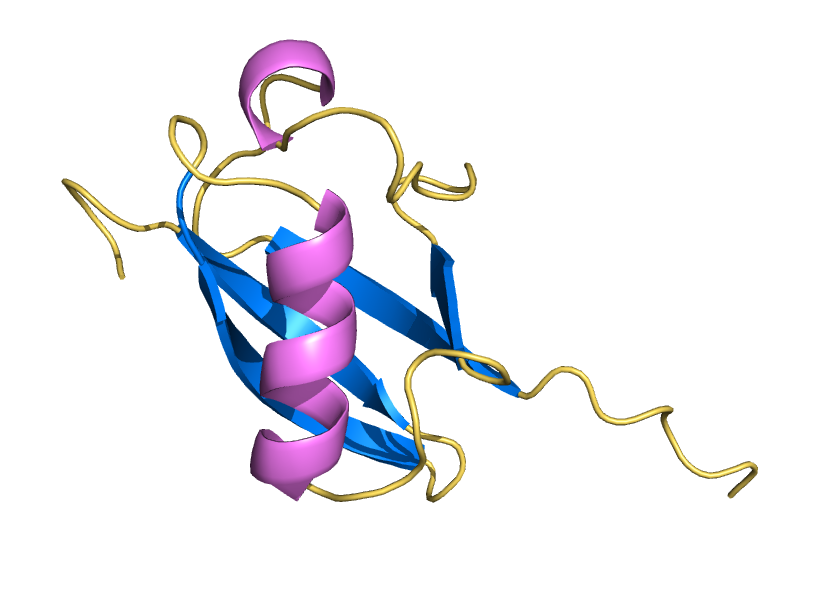
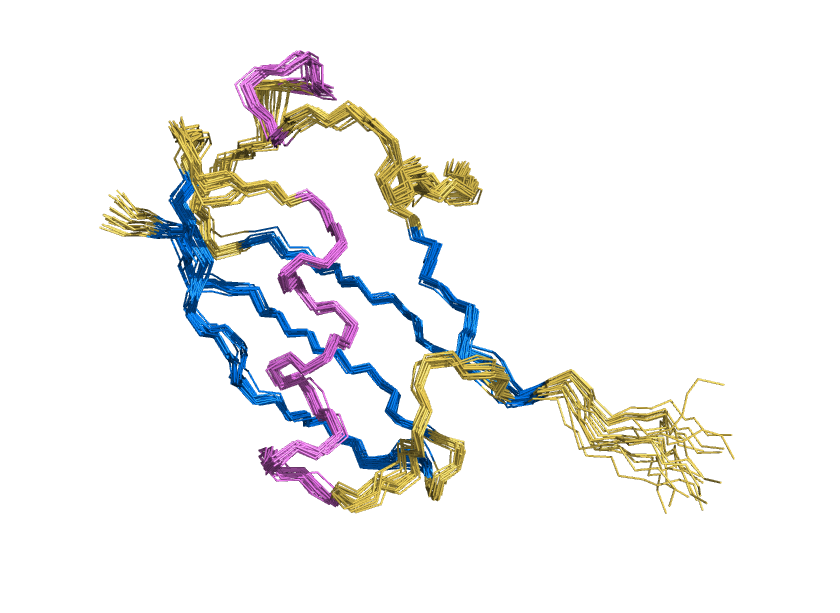
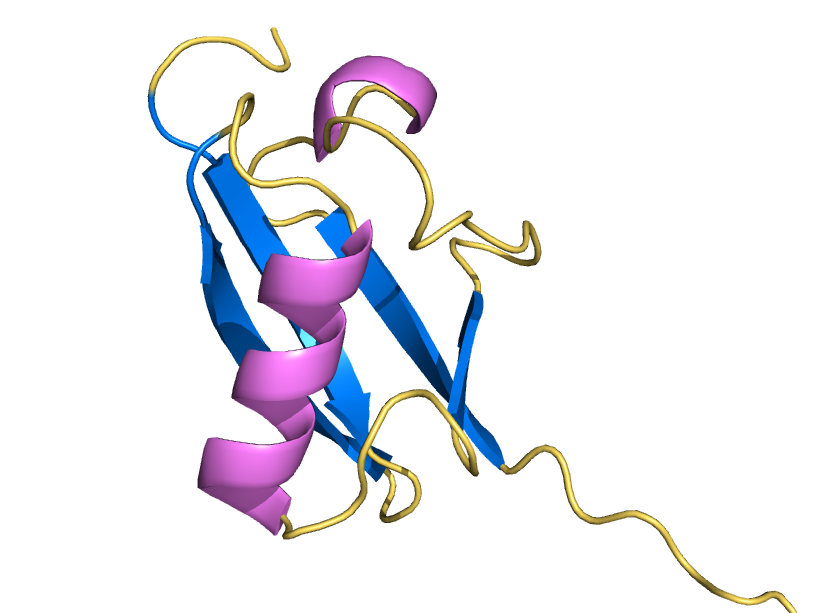
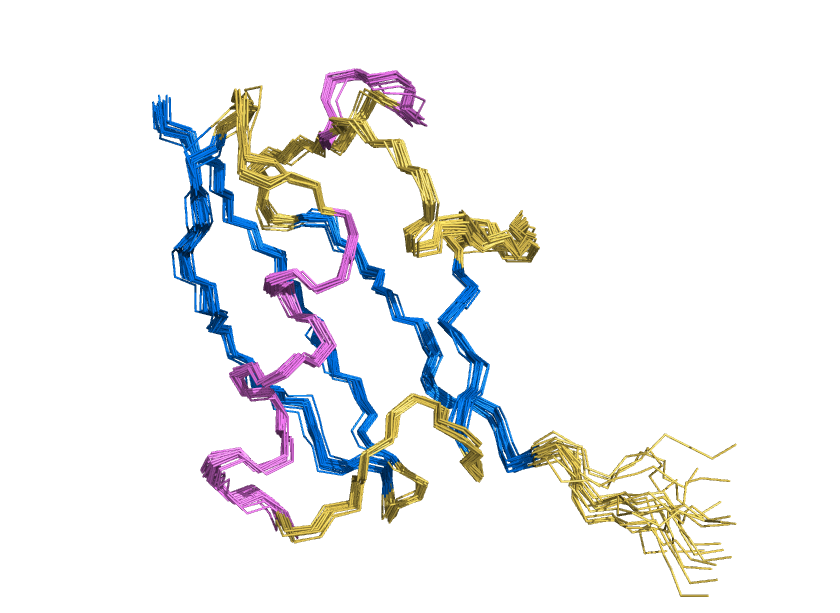
Water shell forms a coat for AI-designed ubiquitin-fold protein to prevent it from denaturation
ProteinMPNN has demonstrated a striking ability to redesign proteins in ways that unexpectedly increase thermal stability. How does this happen? In our study, we found that water is the key player. The redesigned protein surfaces recruit and retain water molecules, forming a structured hydration “shell” that wraps the 3D fold and helps protect it from unfolding. Two examples shown here that ubibquitin variant R4 remains folded under chemical denaturation condition (8 M urea). This work has been published in the Journal of the American Chemical Society.
CryoEM Reveals Mechanism of Polymyxin-resistant PmrA-bound Transcription Activation Complex
The cryoEM structure composed of a dimeric PmrA, double stranded DNA, RNA polymerase and cofactor sigma70 was determined and the work is published in Nucleic Acids Research (2023-Sept).
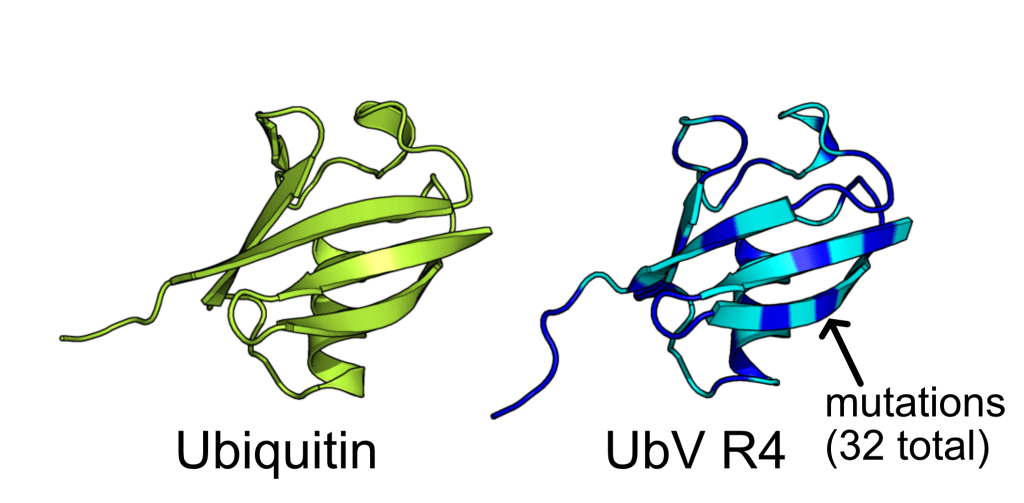
AI-designed Ubiquitin Variant acts as an effective modulator of Rsp5 E3 ligase
We demonstrated the crystal structure of a synthetic ubiquitin variant named R4 (PDB 8J0A) designed by an AI tool ProteinMPNN is identical to wild type ubiquitin. The R4 is an effective allosteric activator of Rsp5 E3 ligase. Work published in ACS Synthetic Biology.
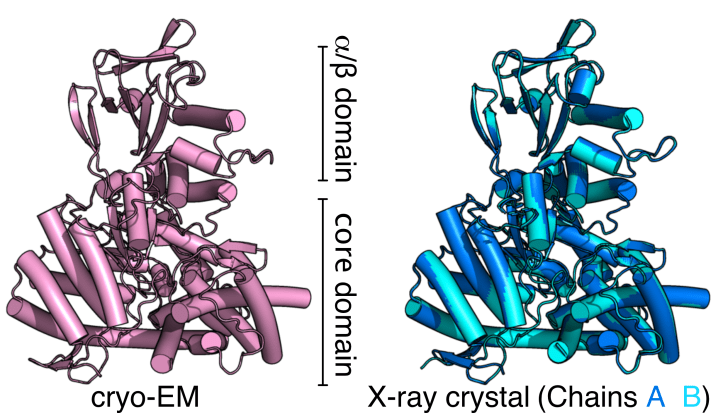
Cryo-EM reveals high-quality structure and conformational dynamics of a 723-aa Malate Synthase G (MSG)
Cryo-EM structure of 723-aa malate synthase G at 2.9 Å (PDB ID 7YQM) and the 1.6 Å X-ray crystal structure of malate synthase G (PDB ID 7YQN), published in Journal of Structural Biology.
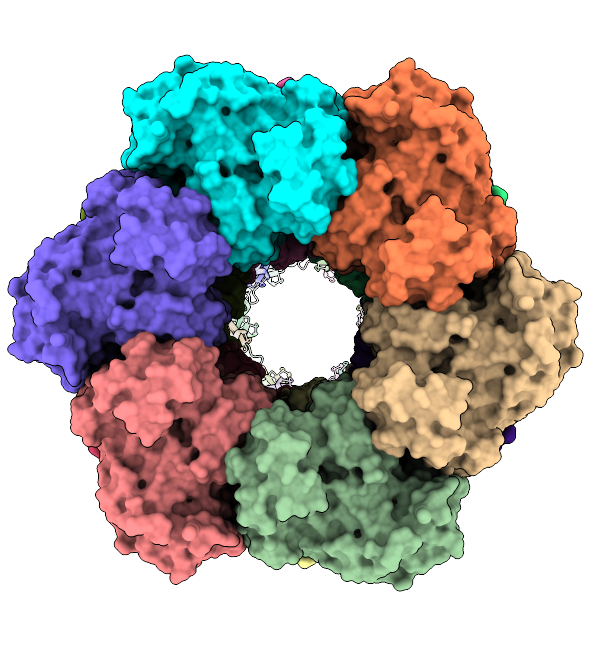
The mysterious filament of Ecoli glutamine synthetase decoded by cryoEM
The assembly of filamentous structure of E coli glutamine synthetase, published in Protein Science. PDB ID = 7W85, EMDB entry = 32352.
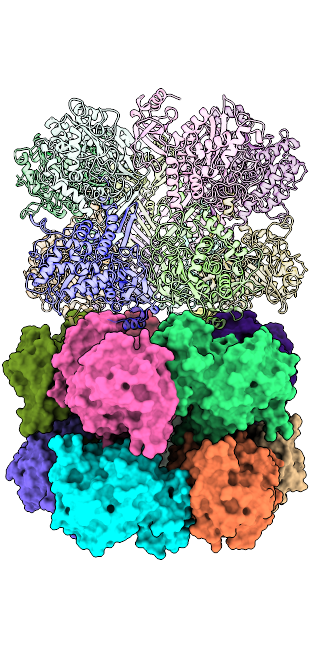
Papain-like protein of SARS-CoV-2 (C111S mutant), PDB ID = 7D47