Above is the image generated by Gemini 3.1 using this post.
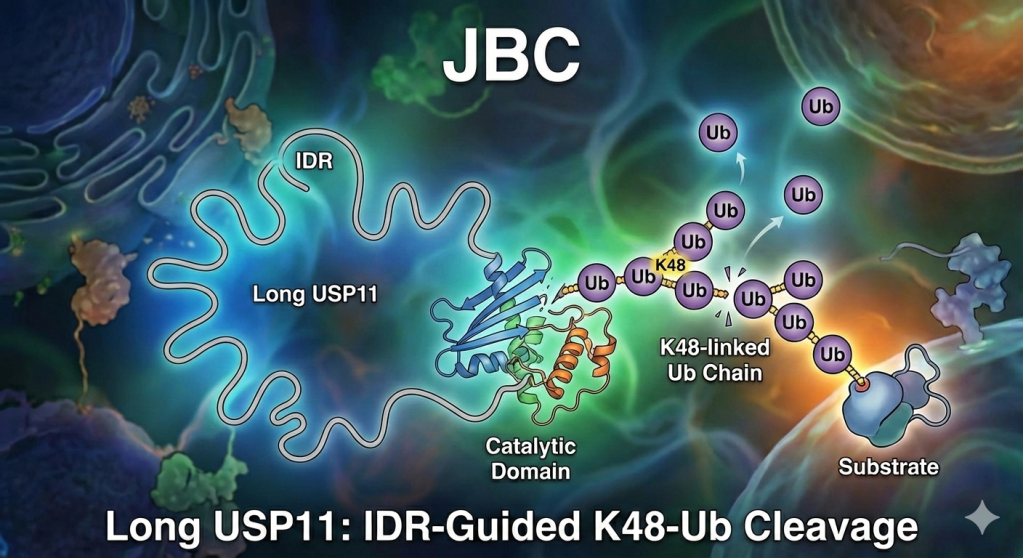
In our recent paper in Journal of Biological Chemistry, we set out to understand how USP11, an enzyme that removes ubiquitin tags from proteins, knows which targets to act on. USP11 is special because its catalytic domain is split in two, with a UBL2 domain and a flexible region (IDR) inserted in between.
By studying tetrameric ubiquitin chains (Ub₄)—a more realistic version of what happens in cells—we found that this inserted UBL2 region gives USP11 a sharp preference for K48-linked chains, which are the signals that mark proteins for degradation. Without UBL2, USP11 loses that precision. Interestingly, its close relatives, USP4 and USP15, don’t rely on this mechanism at all.
We also used AI-driven docking (DiffDock) to look for small molecules that could block USP11. This screen revealed two FDA-approved drugs, Fenoldopam and Olanzapine, as potent inhibitors. They fit neatly into USP11’s S1 ubiquitin-binding site, preventing the enzyme from cutting the ubiquitin tail.
Together, these results show how a non-catalytic domain fine-tunes enzyme selectivity—and how AI-assisted screening can help repurpose known drugs to target disease-related enzymes like USP11.

Below is the automatic generated slides by NotebookLM. The gel images in the slides are not real data. Be caution.