cheating sheet:
lv: call for LACS analysis
st: calibrate nuclei frequency offset
es: generate NMRstar
Generating an NMRstar formatted file with assigned chemical shifts can be efficiently accomplished using the POKY tool. As the NMRstar format is commonly used for data deposition at BMRB, I often perform chemical shift calibration by employing the LACS (Linear Analysis of Chemical Shifts) function. In cases where the calibration is conducted using reference compounds such as DSS and TSP (see my previous post), the LACS analysis provides useful information to assess the accuracy of the chemical shift calibration.
In Poky, with the spectra assigned, call shortcut “lv” to run LACS analysis and visualization. Simply click “Run” button, point the directory to save files.

Once LACS analysis is finished, Poky asks whether user wants to visualize the validation results or not. If No, users will only see output text in the previous pyLACS window.

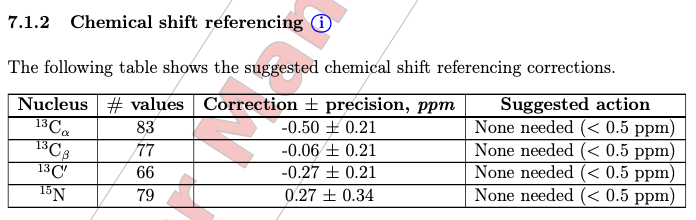
By choose “Yes” button, we can see 3 plots covering CACB-vs-CO/CA/CB correlation. The example using ubiquitin are showing here:

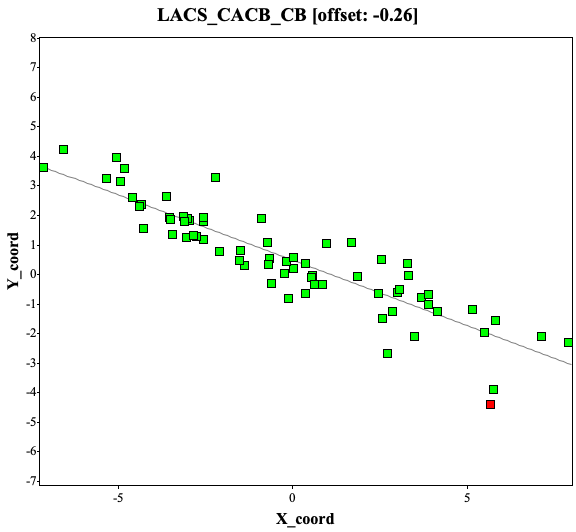

The offsets are -0.26, -0.26, and 0 ppm for 13C-alpha, 13C-beta, and 13C atoms, respectively. The values between -0.5 to 0.5 ppm are considered okay while deposition.
If the chemical shifts are not calibrated well, the LACS analysis will point out the offsets. Here are the examples of CACB_CA and CACB_CB correlation plots, showing a 2.31 ppm offset.
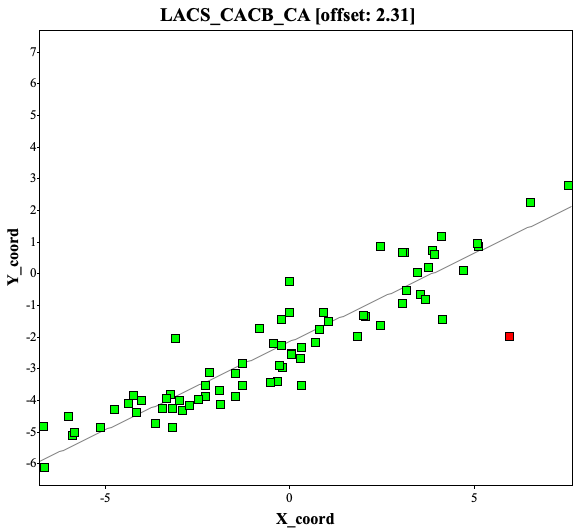
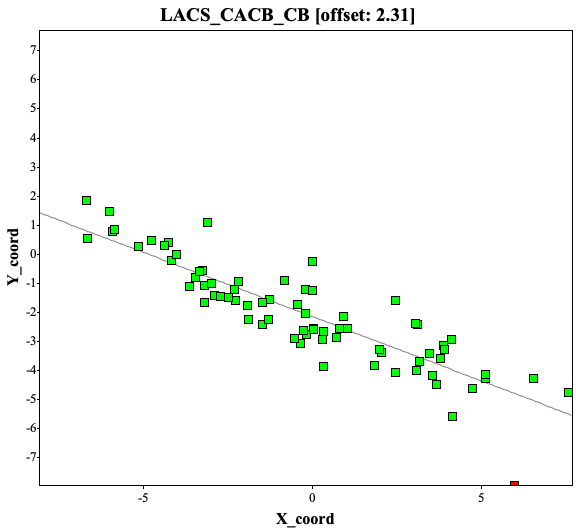
Because of frequency offset, while depositing the assignment to BMRB, user will receive warnings of significant offset (top) and after correction of offset, the report seems okay (bottom, i used different protein assignments at here)
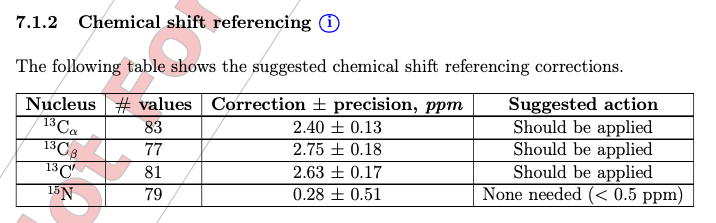
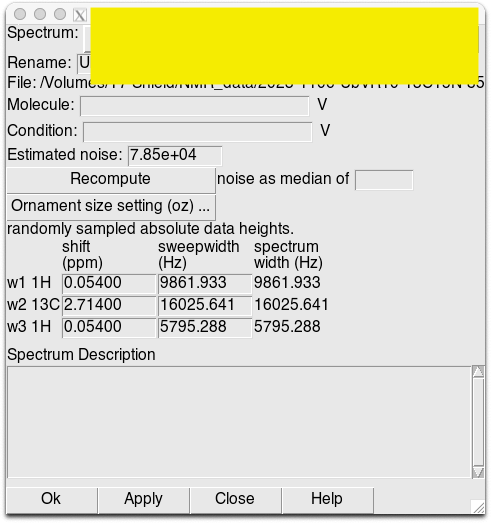
To calibrate the spectra, use “st” function in Poky to calibrate the spectra in each 2D and 3D spectrum.
Once the LACS analysis is good, type “es” shortcut to generate a NMRstar file is straightforward.