In Academia Sinica CryoEM Center (ASCEM), Dr. Yuan-Chih (Kondo) Chang and Dr. Chun-Hsiung Wang used the newly commercial HexAuFoil grid to collect 9,017 movies on our Titan Krios microscope equipped with a K3 detector. My group provided the purified mouse apoferritin as usual.
The HexAuFoil greatly minimized the drift effect compared to QuantiFoil Holey Carbon grid we commonly used. Below are example images of the grid overview and the square overview of this HexAuFoil grid, respectively.

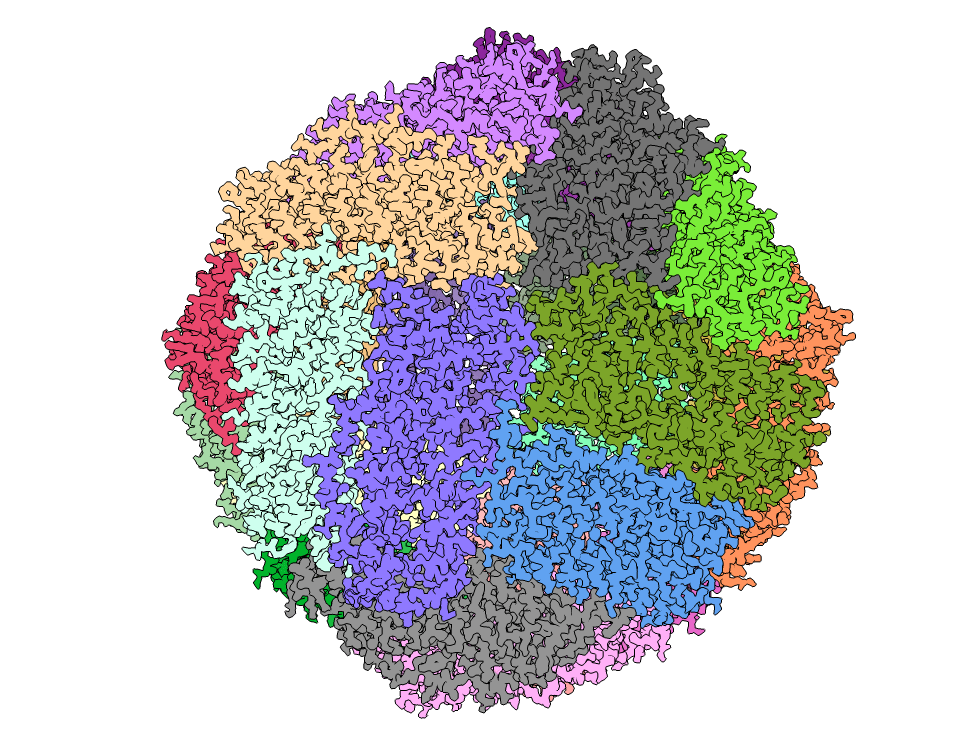
The ASCEM team finally achieved a 1.5 Å map of apoferritin using 1,020,095 particles. The map, built coordinate, and raw data were deposited to EMDB, RCSB-PDB, and EMPIAR, respectively. Accession code can be found in the table below.
| Database | Accession Code |
| RCSB-PDB | 9IUY (RCSB, PDBj, PDBe) |
| EMDB | 60915 (EMDB, PDBj) |
| EMPIAR | 12240 (EMPIAR, EMPIAR @ PDBj) |
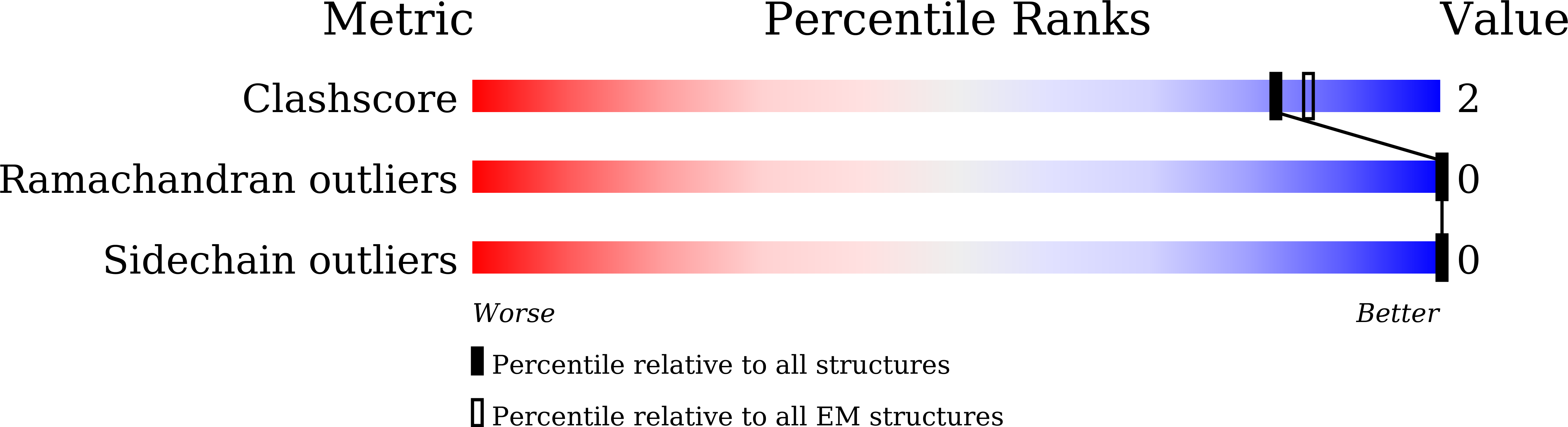
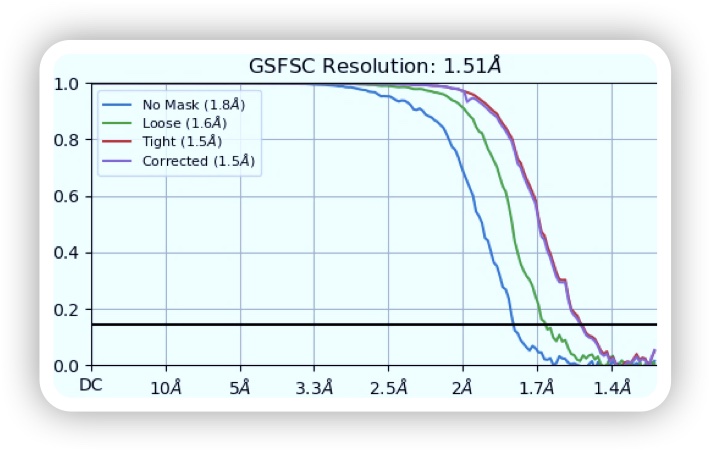
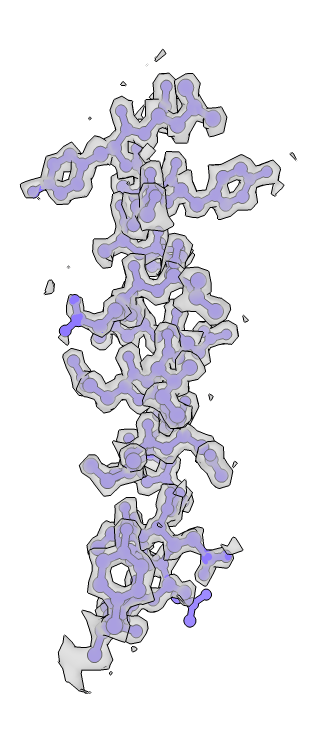
CryoSparc was used to reconstruct the 3D map of apoferritin and structure model was built. The structural quality is great as no outliers were found and the molprobity clashscore is low (see the PDB validation bar graph above). Below images show the map colored without and with structural chains. I also extracted the map covered residues 12-35 of chain A (using ChimeraX). The holes in the benzene rings of three TYR residues are vivid. Sidechains maps and atoms are well matched presenting a cryoEM structure of apoferritin.