There are several tools providing file conversion between Xplor-NIH and Cyana, but I found PdbStat is a pain-free one. Other scripts/tools, in my tests, remain issues sometimes.
PdbStat was developed by Guy Montelione’s group and was published in 2013 in J Biomol NMR. 2013 Aug; 56(4): 337–351 (doi: 10.1007/s10858-013-9753-7): PDBStat: A Universal Restraint Converter and Restraint Analysis Software Package for Protein NMR.
“Provides extensive tools for assessing NMR structures against data, including restraint violation analysis and RDC analysis, as well as tools for converting between coordinate and restraint formats, including NEF format, structural superimposition, and contact map generation.” quoted from PdbStat’s website.
PdbStat runs in Linux and Mac (OS arm64 or old x86_64). It can be downloaded at https://github.rpi.edu/RPIBioinformatics/PDBStat_public. The NESG Wiki for PdbStat is also very informative, useful, and a good reference. Highly recommended.
I use PdbStat to convert Cyana-generated NMR restraints (distance, coupling constants, sequence) to Xplor-NIH format for further calculation. Here are the steps to get the files converted smoothly.
- Execute PdbStat in the X-terminal.
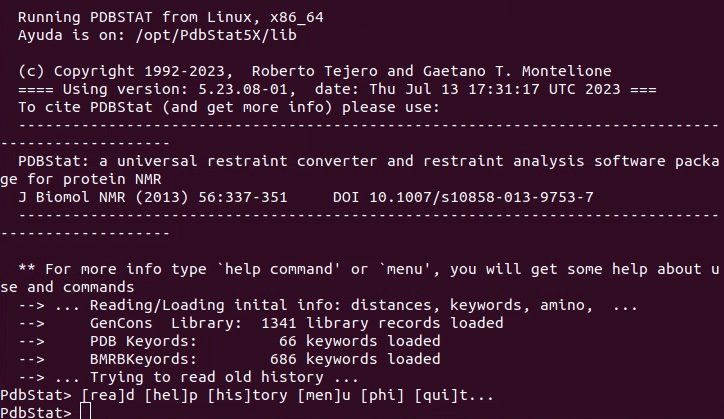
2. Load PDB coordinate or sequence file. This is required for restraint conversion as PdbStat needs to know the residue types and positions. I prefer structure PDB file.
In PdbStat, use “rea (or read)” to read file, use “save” to output file.
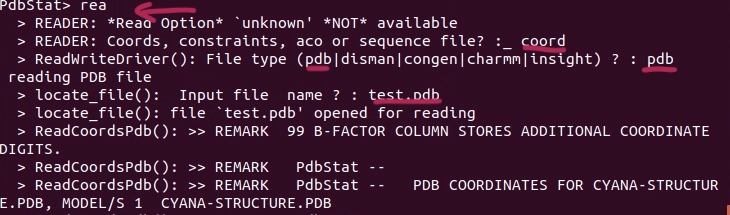
3. Load the Cyana NOE restraints (.upl), remember to choose “cyana” format which is not reminded in the listed options.
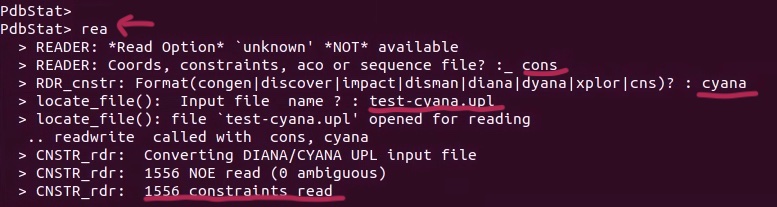
4. Save as Xplor-NIH NOE restraints (i save it as noe.tbl). PdbStat asked to estimate/add the range of upper/lower bound. Users can define the percentages. I often use 10-15%.
5. Load the Cyana TALOS-generated dihedral angle restraints (test.aco)
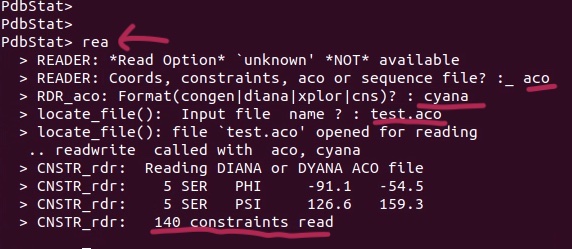
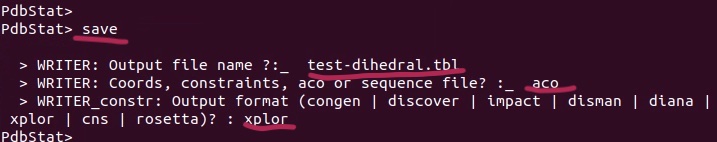
6. Save as Xplor-NIH dihedral angle restraints (I save it as dihedral.tbl).
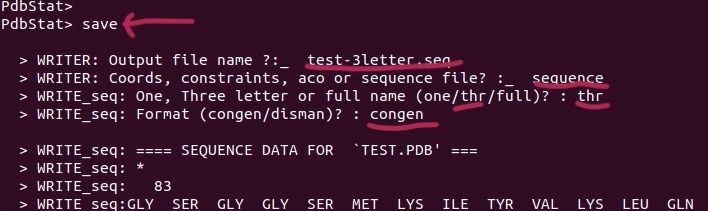
7. Save the sequence in 3-letter code format for Xplor-NIH
Here are the text format of the files
Dihedral angles:
cyana:
5 SER PHI -91.1 -54.5
5 SER PSI 126.6 159.3
6 MET PHI -153.3 -96.8
6 MET PSI 108.9 157.9
7 LYS PHI -129.7 -74.2
7 LYS PSI 118.2 158.6
8 ILE PHI -161.3 -116.5
8 ILE PSI 139.5 173.1
XPLOR-NIH:
assign (resid 4 and name C ) (resid 5 and name N )
(resid 5 and name CA ) (resid 5 and name C ) 1.0 -72.800 18.300 2
assign (resid 5 and name N ) (resid 5 and name CA )
(resid 5 and name C ) (resid 6 and name N ) 1.0 142.950 16.350 2
assign (resid 5 and name C ) (resid 6 and name N )
(resid 6 and name CA ) (resid 6 and name C ) 1.0 -125.050 28.250 2
assign (resid 6 and name N ) (resid 6 and name CA )
(resid 6 and name C ) (resid 7 and name N ) 1.0 133.400 24.500 2
assign (resid 6 and name C ) (resid 7 and name N )
(resid 7 and name CA ) (resid 7 and name C ) 1.0 -101.950 27.750 2
assign (resid 7 and name N ) (resid 7 and name CA )
(resid 7 and name C ) (resid 8 and name N ) 1.0 138.400 20.200 2
assign (resid 7 and name C ) (resid 8 and name N )
(resid 8 and name CA ) (resid 8 and name C ) 1.0 -138.900 22.400 2
assign (resid 8 and name N ) (resid 8 and name CA )
(resid 8 and name C ) (resid 9 and name N ) 1.0 156.300 16.800 2
assign (resid 8 and name C ) (resid 9 and name N )
(resid 9 and name CA ) (resid 9 and name C ) 1.0 -121.000 16.400 2
NOE distances:
Cyana format:
28 ILE HB 57 ASP HA 3.11 #peak 8 #plist 1 #SUP 0.87 #QF 0.87
66 ILE HB 66 ILE QD1 3.23 #peak 12 #plist 1 #SUP 0.95 #QF 0.95
23 GLU HA 24 PRO HD2 3.28 #peak 18 #plist 1 #SUP 0.98 #QF 0.98
55 LEU HA 55 LEU QD2 2.90 #peak 20 #plist 1 #SUP 0.90 #QF 0.90
53 LYS QB 53 LYS QG 2.77 #peak 22 #plist 1 #SUP 0.99 #QF 0.93
31 VAL HA 31 VAL QG1 3.05 #peak 25 #plist 1 #SUP 0.99 #QF 0.29
Xplor-NIH format:
assign (resid 28 and name HB ) (resid 57 and name HA ) 2.701 0.901 0.720
! target: 2.70 lo: 1.80 up: 3.42 [1, idx: 401 895]
assign (resid 66 and name HB ) (resid 66 and name HD1*) 2.766 0.966 0.787
! target: 2.77 lo: 1.80 up: 3.55 [1, idx: 1032 0]
assign (resid 23 and name HA ) (resid 24 and name HD2 ) 2.794 0.994 0.814
! target: 2.79 lo: 1.80 up: 3.61 [1, idx: 333 0]
assign (resid 55 and name HA ) (resid 55 and name HD2*) 2.585 0.785 0.605
! target: 2.58 lo: 1.80 up: 3.19 [1, idx: 861 0]
assign (resid 53 and name HB* ) (resid 53 and name HG* ) 2.513 0.713 0.534
! target: 2.51 lo: 1.80 up: 3.05 [1, idx: 825 828]
assign (resid 31 and name HA ) (resid 31 and name HG1*) 2.668 0.868 0.688
! target: 2.67 lo: 1.80 up: 3.36 [1, idx: 452 0]