In the NSRRC protein crystal diffraction beam line station, users can use the licensed HKL2000 to spot the diffractions and index/scale the refined space group. This a convenient service provided NSRRC but it takes time to manual process. Moreover, it is a bit inconvenient for remote access users to operate data collection (BlueICE) and data process (HKL2000) at the same time.
I used to use XDS to run semi-automatic data process for the diffraction data and recently learned xia2+DIALS provided in CCP4 can perform fully automatic data process. This is a very convenient tool to quickly judge the quality of collected data. If needed, users can refine the strategy to collect more images with a smaller oscillation, increase the exposure time, or perform helical data collection.


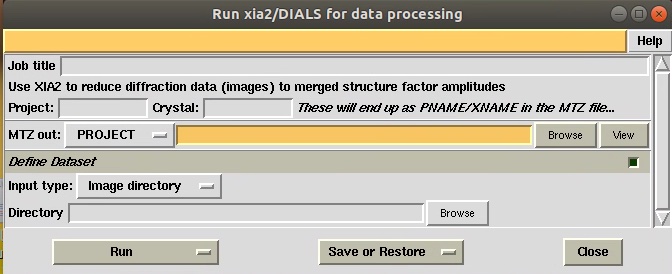
Users can run XIA2 through CCP4 or simply use the command line mode when an “xinfo” profile is prepared. Below is the simplified GUI for xia2. Just define the local of the diffraction images (in NSRRC, we define the directory and xia2 will deal with the master.h5 and data.h5 file automatically) and click “Run”. It often takes 10-30 minutes to perform indexing, integrating and scaling depending on the numbers of images, data quality and computational resources.
I like to run Xia2 in the the terminal as I can specify some arguments for data process. Here are the steps I used to use:
1. Edit an xinfo file
2. Execute xia in the terminal without specified arguments
3. Repeat #2 but specify the highest shell quality with I/sigma = 0.25
The xinfo file can be something like below (save as automatic.xinfo). Be sure about the wavelength, directory, image path/name, and the start/end image.
BEGIN PROJECT AUTOMATIC
BEGIN CRYSTAL DEFAULT
BEGIN WAVELENGTH NATIVE
WAVELENGTH 0.976246
END WAVELENGTH NATIVE
BEGIN SWEEP SWEEP1
WAVELENGTH NATIVE
DIRECTORY /ssd/xia/kpw005/P10
IMAGE kpw005_15_0001_master.h5
START_END 1 180
END SWEEP SWEEP1
END CRYSTAL DEFAULT
END PROJECT AUTOMATIC
Then I ran xia in terminal like:
xia2 xinfo=automatic.xinfo
or
xia2 xinfo=automatic.xinfo cc_half=none misigma=1 isigma=0.25
To run the two commands, I duplicate the working folder and run xia2 (latter command) in the duplicated folder to avoid overwriting files.
The first command “xia2 xinfo=automatic.info” will use the highest diffraction shell by default and trim the data with a threshold of CC_half = 0.3. Once the xia2 job is finished, it generated a “xia2.txt” output which is very useful for judging the data quality (and future publication). The xia2.txt of the first command is shown below. The highest shell is 1.47 – 1.49 Å.
Environment configuration...
Python => /opt/ccp4/ccp4-8.0/libexec/python3.7
CCTBX => /opt/ccp4/ccp4-8.0/lib/python3.7/site-packages
CCP4 => /opt/ccp4/ccp4-8.0
CCP4_SCR => /tmp/tmpthlj60gf
Starting directory: /ssd/xia/xia2_P10_1
Working directory: /ssd/xia/xia2_P10_1
Free space: 3026.91 GB
Host: xxx
Contact: xia2.support@gmail.com
XIA2 3.8.6
DIALS 3.8
CCP4 8.0.017
Command line: xia2 xinfo=automatic.xinfo
Project directory: /ssd/xia/xia2_P10_1
-------------------- Spotfinding SWEEP1 --------------------
50736 spots found on 180 images (max 1131 / bin)
*
* * *
* ************************* * *
************************************************* ** * * * *
************************************************************
************************************************************
************************************************************
************************************************************
************************************************************
************************************************************
1 image 180
------------------- Autoindexing SWEEP1 --------------------
All possible indexing solutions:
oI 57.58 62.76 89.76 90.00 90.00 90.00
mC 85.16 89.75 57.57 90.00 132.53 90.00
aP 57.57 61.87 61.87 93.00 117.73 117.73
Indexing solution:
oI 57.58 62.76 89.76 90.00 90.00 90.00
-------------------- Integrating SWEEP1 --------------------
Processed batches 2 to 181
Standard Deviation in pixel range: 0.38 0.56
Integration status per image (60/record):
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
"o" => good "%" => ok "!" => bad rmsd
"O" => overloaded "#" => many bad "." => weak
"@" => abandoned
Mosaic spread: 0.731 < 0.731 < 0.731
-------------------- Preparing DEFAULT ---------------------
--------------------- Scaling DEFAULT ----------------------
Resolution for sweep NATIVE/SWEEP1: 1.47 (cc_half > 0.3)
--------------------- Scaling DEFAULT ----------------------
---------------- Systematic absences check -----------------
Most likely space group: I 2 2 2
------------------- Unit cell refinement -------------------
Overall: 57.55 62.73 89.73 90.00 90.00 90.00
Project: AUTOMATIC
Crystal: DEFAULT
Sequence:
Wavelength name: NATIVE
Wavelength 0.97625
Sweeps:
SWEEP SWEEP1 [WAVELENGTH NATIVE]
TEMPLATE kpw005-P10_15_0001_master.h5
DIRECTORY /ssd/xia/kpw005/P10
IMAGES (USER) 1 to 180
MTZ file: /ssd/xia/xia2_P10_1/DEFAULT/NATIVE/SWEEP1/integrate/12_integrated.refl
For AUTOMATIC/DEFAULT/NATIVE Overall Low High
High resolution limit 1.47 3.98 1.47
Low resolution limit 28.78 28.78 1.49
Completeness 96.1 100.0 68.7
Multiplicity 5.2 6.0 2.1
I/sigma 5.1 27.3 0.1
Rmerge(I) 0.118 0.068 2.168
Rmerge(I+/-) 0.108 0.064 2.349
Rmeas(I) 0.130 0.074 2.766
Rmeas(I+/-) 0.129 0.076 3.088
Rpim(I) 0.052 0.031 1.684
Rpim(I+/-) 0.069 0.041 1.982
CC half 0.991 0.989 0.363
Wilson B factor 15.680
Anomalous completeness 88.0 100.0 37.9
Anomalous multiplicity 2.9 3.4 1.4
Anomalous correlation -0.173 -0.147 0.037
Anomalous slope 0.260
dF/F 0.104
dI/s(dI) 0.356
Total observations 140788 9170 1920
Total unique 26956 1523 931
Assuming spacegroup: I 2 2 2
Unit cell (with estimated std devs):
57.5512(6) 62.7304(6) 89.7337(7)
90.0 90.0 90.0
mtz_unmerged format:
Scaled reflections (NATIVE): /ssd/xia/xia2_P10_1/DataFiles/AUTOMATIC_DEFAULT_scaled_unmerged.mtz
mtz format:
Scaled reflections: /ssd/xia/xia2_P10_1/DataFiles/AUTOMATIC_DEFAULT_free.mtz
Processing took 00h 06m 50s
XIA2 used... ccp4, dials, dials.scale, xia2
Here are the appropriate citations (BIBTeX in xia2-citations.bib.)
Beilsten-Edmands, J. et al. (2020) Acta Cryst. D76.
Winn, M. D. et al. (2011) Acta Cryst. D67, 235-242.
Winter, G. (2010) J. Appl. Cryst. 43, 186-190.
Winter, G. et al. (2018) Acta Cryst. D74, 85-97.
Status: normal termination
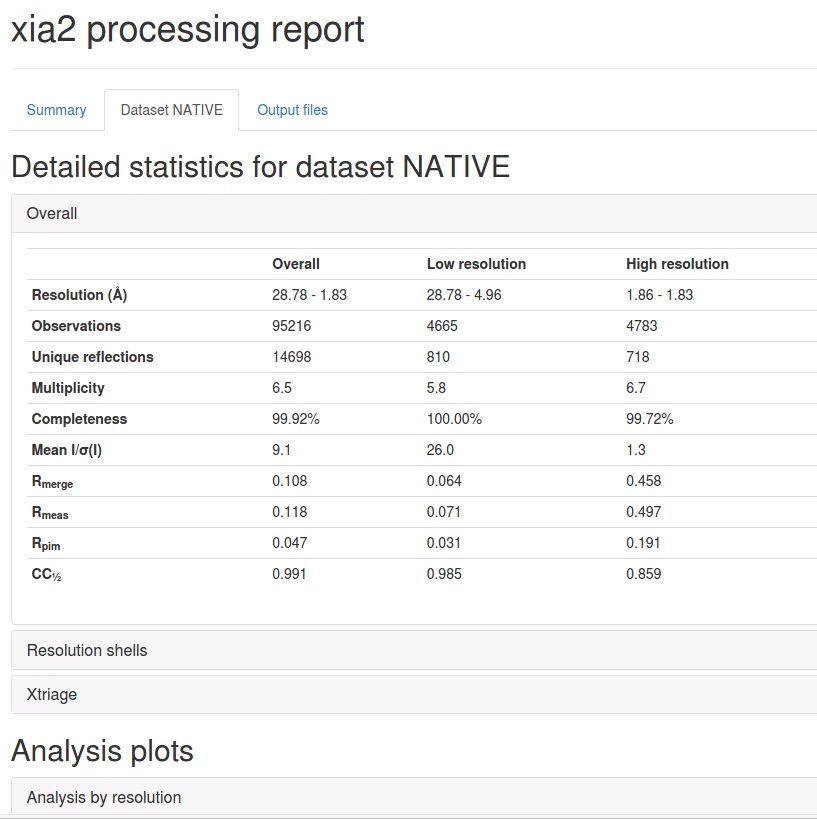
For the second command, with a specified cut-off, I can quickly estimate the best/reasonable highest diffraction shell to be used for molecular replacement and following model building. The xia2.txt with specified merged I/sigma range is shown below. This time, the highest shell changed from 1.47-1.49 Å to 1.83-1.86 Å. The new one retains better data quality for the high-resolution shells compared to the default one (no specification).
Environment configuration...
Python => /opt/ccp4/ccp4-8.0/libexec/python3.7
CCTBX => /opt/ccp4/ccp4-8.0/lib/python3.7/site-packages
CCP4 => /opt/ccp4/ccp4-8.0
CCP4_SCR => /tmp/tmpssdrjgct
Starting directory: /ssd/xia/xia2_P10_2
Working directory: /ssd/xia/xia2_P10_2
Free space: 3025.67 GB
Host: xxxx
Contact: xia2.support@gmail.com
XIA2 3.8.6
DIALS 3.8
CCP4 8.0.017
Command line: xia2 xinfo=automatic.xinfo cc_half=none misigma=1 isigma=0.25
Project directory: /ssd/xia/xia2_P10_2
-------------------- Spotfinding SWEEP1 --------------------
50736 spots found on 180 images (max 1131 / bin)
*
* * *
* ************************* * *
************************************************* ** * * * *
************************************************************
************************************************************
************************************************************
************************************************************
************************************************************
************************************************************
1 image 180
------------------- Autoindexing SWEEP1 --------------------
All possible indexing solutions:
oI 57.58 62.76 89.76 90.00 90.00 90.00
mC 85.16 89.75 57.57 90.00 132.53 90.00
aP 57.57 61.87 61.87 93.00 117.73 117.73
Indexing solution:
oI 57.58 62.76 89.76 90.00 90.00 90.00
-------------------- Integrating SWEEP1 --------------------
Processed batches 2 to 181
Standard Deviation in pixel range: 0.38 0.56
Integration status per image (60/record):
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
oooooooooooooooooooooooooooooooooooooooooooooooooooooooooooo
"o" => good "%" => ok "!" => bad rmsd
"O" => overloaded "#" => many bad "." => weak
"@" => abandoned
Mosaic spread: 0.731 < 0.731 < 0.731
-------------------- Preparing DEFAULT ---------------------
--------------------- Scaling DEFAULT ----------------------
Resolution for sweep NATIVE/SWEEP1: 1.83 (merged <I/sigI> > 1.0)
--------------------- Scaling DEFAULT ----------------------
---------------- Systematic absences check -----------------
Most likely space group: I 2 2 2
------------------- Unit cell refinement -------------------
Overall: 57.55 62.73 89.73 90.00 90.00 90.00
Project: AUTOMATIC
Crystal: DEFAULT
Sequence:
Wavelength name: NATIVE
Wavelength 0.97625
Sweeps:
SWEEP SWEEP1 [WAVELENGTH NATIVE]
TEMPLATE kpw005-P10_15_0001_master.h5
DIRECTORY /ssd/xia/kpw005/P10
IMAGES (USER) 1 to 180
MTZ file: /ssd/xia/xia2_P10_2/DEFAULT/NATIVE/SWEEP1/integrate/12_integrated.refl
For AUTOMATIC/DEFAULT/NATIVE Overall Low High
High resolution limit 1.83 4.96 1.83
Low resolution limit 28.78 28.78 1.86
Completeness 99.9 100.0 99.7
Multiplicity 6.5 5.8 6.7
I/sigma 9.1 26.0 1.3
Rmerge(I) 0.108 0.064 0.458
Rmerge(I+/-) 0.100 0.061 0.410
Rmeas(I) 0.118 0.071 0.497
Rmeas(I+/-) 0.119 0.074 0.483
Rpim(I) 0.047 0.031 0.191
Rpim(I+/-) 0.064 0.041 0.253
CC half 0.991 0.985 0.859
Wilson B factor 20.540
Anomalous completeness 99.6 100.0 99.7
Anomalous multiplicity 3.4 3.3 3.5
Anomalous correlation -0.238 -0.372 -0.134
Anomalous slope 0.402
dF/F 0.088
dI/s(dI) 0.542
Total observations 95216 4665 4783
Total unique 14698 810 718
Assuming spacegroup: I 2 2 2
Unit cell (with estimated std devs):
57.5512(6) 62.7304(6) 89.7337(7)
90.0 90.0 90.0
mtz_unmerged format:
Scaled reflections (NATIVE): /ssd/xia/xia2_P10_2/DataFiles/AUTOMATIC_DEFAULT_scaled_unmerged.mtz
mtz format:
Scaled reflections: /ssd/xia/xia2_P10_2/DataFiles/AUTOMATIC_DEFAULT_free.mtz
Processing took 00h 06m 41s
XIA2 used... ccp4, dials, dials.scale, xia2
Here are the appropriate citations (BIBTeX in xia2-citations.bib.)
Beilsten-Edmands, J. et al. (2020) Acta Cryst. D76.
Winn, M. D. et al. (2011) Acta Cryst. D67, 235-242.
Winter, G. (2010) J. Appl. Cryst. 43, 186-190.
Winter, G. et al. (2018) Acta Cryst. D74, 85-97.
Status: normal termination
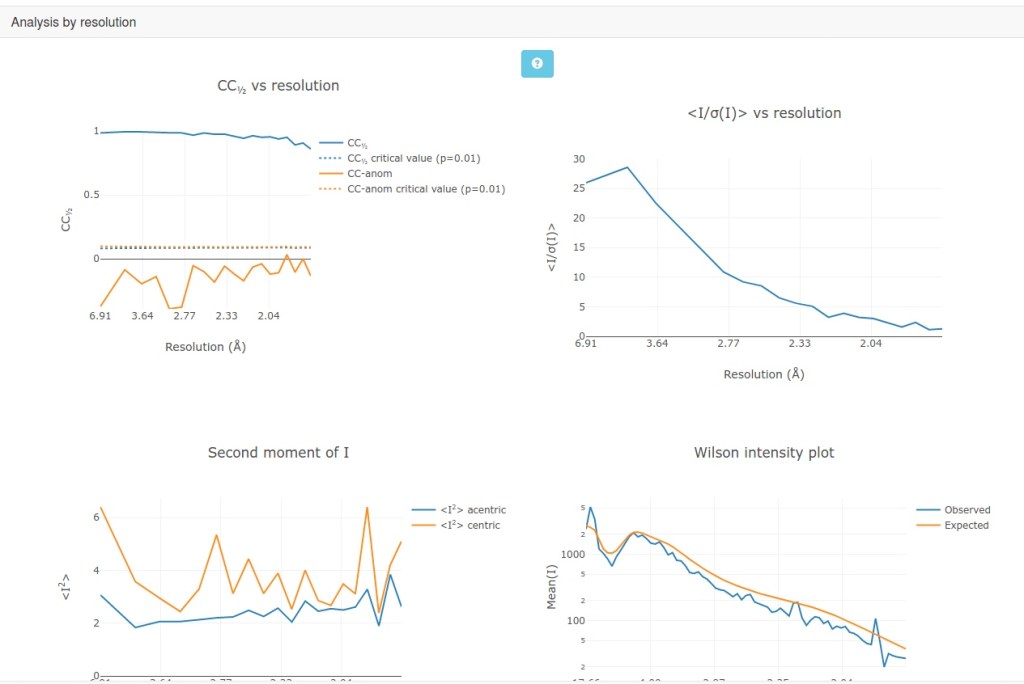
In addition to the text output, the xia2.html provide more analysis for users to know the quality of data. This integrated page is more convenient than sparse information provided in HKL2000 or XDS. Some snapshots are shown below.