We are thrilled to share the exciting news that our work on the Klebsiella pneumoniae PmrA-regulated transcription activation complex has been accepted and published in Nucleic Acids Research on September 4, 2023.
Initially, Dr. Chinpan Chen and Dr. Yuan-Chao Lou from the Institute of Biomedical Sciences proposed this collaboration to me in early 2018. We worked diligently to optimize the expression of RNA polymerase, PmrA, and the DNA bubble sequence. In addition to the previous PmrA-DNA complex structure determined by X-ray crystallography and solution NMR spectroscopy in the Chen group, I designed the cryo-EM experiment, and my group worked on EM map reconstruction and model building.

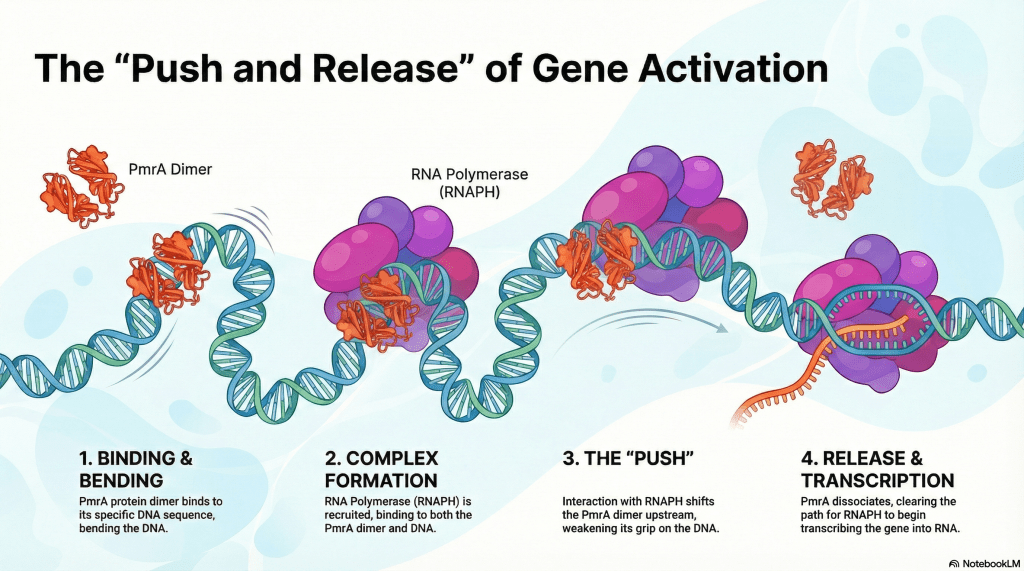
The cryo-EM structure (PDB ID: 8JO2) of the PmrA-regulated transcription activation complex is composed of RNA polymerase core subunits, sigma70, the double-stranded bubble DNA sequence, and the dimeric PmrA. We collected more than 10,000 cryo-EM images with millions of particles. The PmrA-associated RNAPH (RNA polymerase holoenzyme) is transient. We encountered challenges in dealing with the heterogeneous conformation in early 2019, but eventually, the focused map combined with 3D classification revealed a clear picture of the PmrA-bound complex.

Our work reveals a striking DNA shift, indicating that the PmrA binding sequence differs from the previously defined CCTAAT. The cryo-EM data redefines PmrA-DNA interactions when the transcription activation complex is regulated by PmrA, likely representing an escape mode. Since PmrA is linked to polymyxin resistance, we hope that this structural finding opens up opportunities to develop drugs against Klebsiella pneumoniae.