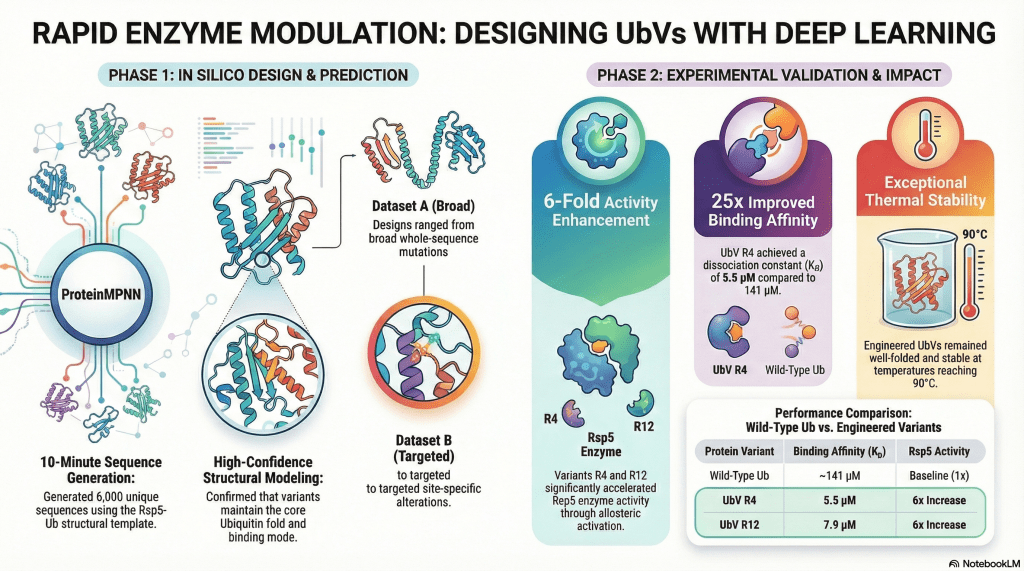
We are very happy to receive a notice email from ACS Synthetic Biology that our manuscript was accepted and online published ahed. The work entitled “Robust design of effective allosteric activators for Rsp5 E3 ligase using the machine-learning tool ProteinMPNN” to show a simple but robust approach to converting ubiquitin into an effective allosteric activator. We used a program called ProteinMPNN right after it was released to the public (GitHub, paper). The goal is to design a new ubiquitin variant (UbV) functioning as a specific activator for the Rsp5 E3 ligase.
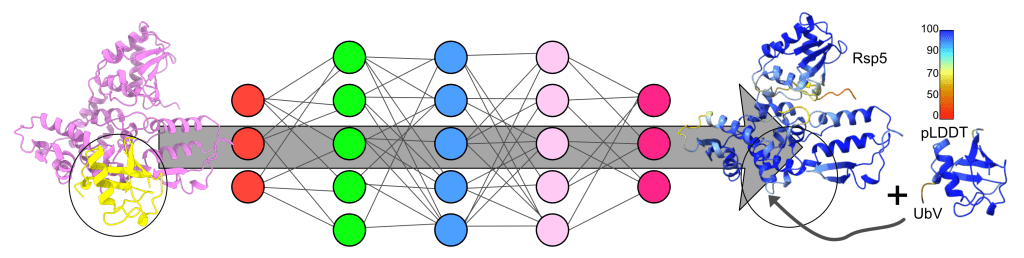
The AI-designed enhanced protein-protein interactions were output as thousand sequences in 10 minutes. Complex structures were first validated by AlphaFold2 prediction revealing promising accuracy. Followed by a series of experiments including (1) oligomeric state using SEC-MALS, (2) secondary structure characterization by circular dichroism (CD), (3) thermal stability by ThermoFluor and CD, (4) crystal structure of R4 (one of the designed UbV), (5) biophysical affinity and (6) activity of Rsp5, we confirmed 4 UbV proteins are effective allosteric activators. Out of 20 synthesized de novo-designed genes, the successful rate is 25%. We believe it can be greatly improved with another design based on the Rsp5-R4 structure. All experiments were finished in 4 months, including a 3-week waiting for gene synthesis. Very efficient. It is very very encouraging, as structural biologists can now design new proteins without the expensive screening/display platform setup. A pure virtual screening makes a magic hit!

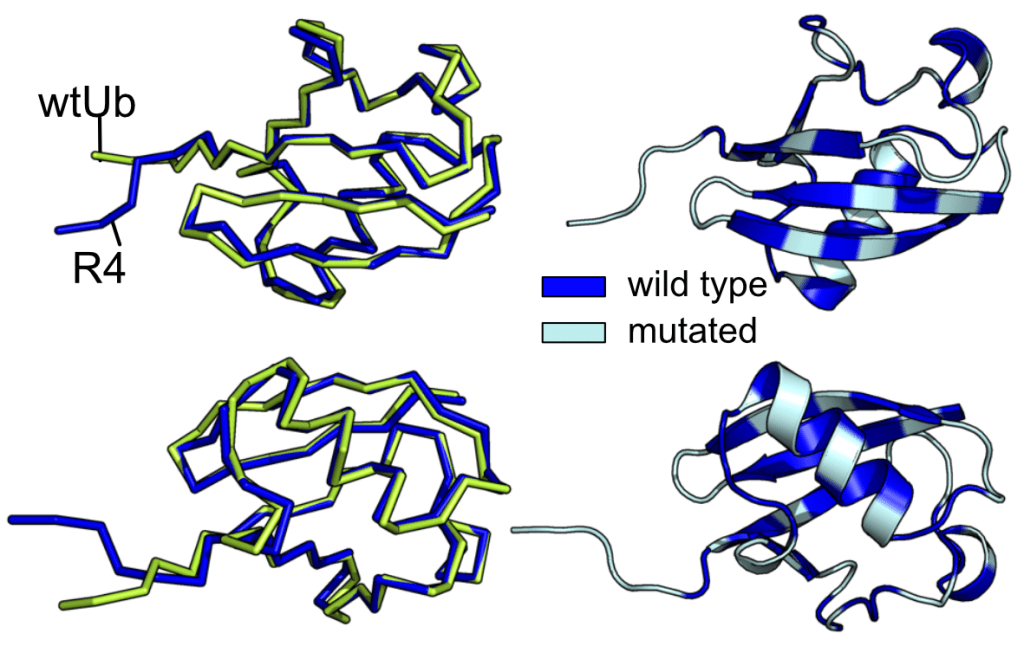
It is worth noting that the R4 crystal structure reveals no difference compared to ubiquitin, even though 32 residues are mutated, and many of them are located in the beta sheets. The PDB ID is 8J0A. ProteinMPNN is a powerful program opening a pandora box of protein design which needed to be feasibly done using an experimental screening approach. The future version of ProteinMPNN or a newcomer AI software will completely change the protein design workflow. Structural biology and biophysics will be very critical to provide experimental validation of the designed function. It is getting more and more important.
The work was primary done hardworking assistants Hsi-Wen and Wei-Lin. We are very thankful for the exceptional Academia Sinica (AS) facilities including AS biophysics core facility (BCF) and AS grid computing center (ASGC) as well as IBC protein core facility (X-ray crystal and biophysics) and National Synchrotron Radiation Research Center (NSRRC).
Below are the summary slides generated by NotebookLM. Note, the structures and gels are somehow hallucinated, but content is okay.